Optimized Deep Neural Network for High-Precision Psoriasis Classification from Dermoscopic Images
DOI:
https://doi.org/10.51252/rcsi.v5i2.996Keywords:
skin disease, long short term memory, psoriasis, efficient netAbstract
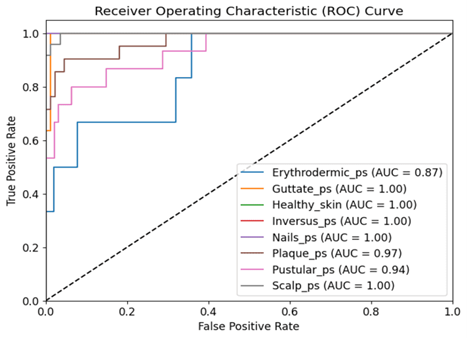
Accurate classification of psoriasis is critical in dermatological diagnostics due to the disease’s diverse clinical presentations and varying severity levels. With numerous subtypes and their visual similarities to other dermatological conditions, precise diagnosis typically requires expert medical knowledge. Early and accurate identification of psoriasis subtypes is essential for initiating timely treatment. This study introduces a novel hybrid deep learning architecture that integrates EfficientNet with Long Short-Term Memory (LSTM) networks for the automated classification of psoriasis from dermoscopic images. The proposed model is designed to simultaneously capture spatial features through EfficientNet and temporal or sequential patterns via LSTM units, thereby improving classification performance. The models are trained and tested on a publicly benchmark dataset comprising 7 distinct classes using the publically available benchmark dataset by Dermnet and BFL-NTU. Experimental results demonstrate that the proposed architecture significantly outperforms the baseline models such as VGG16 and ResNet50, with superior accuracy 89.7% and robust performance across metrics like recall, F1-score with 88%, and Region of Convergence (ROC) of 97%. This compact design with low trainable parameters reduces the computational time and memory makes the model well-suited for deployment for portable devices and enabling real-time mobile-based dermatological assessments.
Downloads
References
Abbas, M., Arslan, M., Yousaf, F., & Khan, A. A. (2024). Enhanced Skin Disease Diagnosis through Convolutional Neural Networks and Data Augmentation Techniques. Journal of Computing & Biomedical Informatics, 7(1). https://doi.org/http://dx.doi.org/10.56979/701/2024
Adegun, A. A., & Viriri, S. (2020). FCN-Based DenseNet Framework for Automated Detection and Classification of Skin Lesions in Dermoscopy Images. IEEE Access, 8, 150377–150396. https://doi.org/10.1109/ACCESS.2020.3016651
Ahmmed, F., Zaheer Raihan, Kamnur Nahar, Asadujjaman, D. M., Mahfujur Rahman, & Abdullah Tamim. (2025). Skin Disease Detection and Classification of Actinic Keratosis and Psoriasis Utilizing Deep Transfer Learning. ArXiv Preprint. https://doi.org/https://doi.org/10.48550/arXiv.2501.13713
Aijaz, S. F., Khan, S. J., Azim, F., Shakeel, C. S., & Hassan, U. (2022). Deep Learning Application for Effective Classification of Different Types of Psoriasis. Journal of Healthcare Engineering, 2022, 1–12. https://doi.org/10.1155/2022/7541583
Aishwarya, U., Daniel, I. J., & Raghul, R. (2020). Convolutional Neural Network based Skin Lesion Classification and Identification. 2020 International Conference on Inventive Computation Technologies (ICICT), 264–270. https://doi.org/10.1109/ICICT48043.2020.9112485
Ayad, H., & Ismail, T. (2020). An Efficient Detection Framework for Linear Skin Lesions with Pigmentary Disorders. 2020 30th International Conference on Computer Theory and Applications (ICCTA), 128–133. https://doi.org/10.1109/ICCTA52020.2020.9477657
Azam, S. N. Z. N., Zakaria, N. H., Hassan, R., & Zulkifle, F. A. (2022). Classification of Psoriasis Microarray Data using Machine Learning. 2022 2nd International Conference on Intelligent Cybernetics Technology & Applications (ICICyTA), 245–249. https://doi.org/10.1109/ICICyTA57421.2022.10038180
Bolia, C., & Joshi, S. (2024). A Comparative Study of Convolutional Neural Network Architecture for Efficient Classification of Psoriasis Disease (pp. 157–171). https://doi.org/10.1007/978-981-97-4533-3_12
DermNetNZ website. (2022). Image Library. https://dermnetnz.org/image-library
Esteva, A., Kuprel, B., Novoa, R. A., Ko, J., Swetter, S. M., Blau, H. M., & Thrun, S. (2017). Dermatologist-level classification of skin cancer with deep neural networks. Nature, 542(7639), 115–118. https://doi.org/10.1038/nature21056
Graves, A., & Schmidhuber, J. (2005). Framewise phoneme classification with bidirectional LSTM and other neural network architectures. Neural Networks, 18(5–6), 602–610. https://doi.org/10.1016/j.neunet.2005.06.042
Greff, K., Srivastava, R. K., Koutnik, J., Steunebrink, B. R., & Schmidhuber, J. (2017). LSTM: A Search Space Odyssey. IEEE Transactions on Neural Networks and Learning Systems, 28(10), 2222–2232. https://doi.org/10.1109/TNNLS.2016.2582924
Griffiths, C. E., & Barker, J. N. (2007). Pathogenesis and clinical features of psoriasis. The Lancet, 370(9583), 263–271. https://doi.org/10.1016/S0140-6736(07)61128-3
Hammad, M., Pławiak, P., ElAffendi, M., El-Latif, A. A. A., & Latif, A. A. A. (2023). Enhanced Deep Learning Approach for Accurate Eczema and Psoriasis Skin Detection. Sensors, 23(16), 7295. https://doi.org/10.3390/s23167295
Huang, C., Yu, A., Wang, Y., & He, H. (2020). Skin Lesion Segmentation Based on Mask R-CNN. 2020 International Conference on Virtual Reality and Visualization (ICVRV), 63–67. https://doi.org/10.1109/ICVRV51359.2020.00024
Jagannathan, V., Priya, R., Sundararajan, V., & Banu, M. (2024). CNN–BiGRU-based hybrid deep learning model for dermatological disease classification. https://doi.org/https://doi.org/10.1016/j.jbi.2024.104006
Krizhevsky, A., Sutskever, I., & Hinton, G. E. (2017). ImageNet classification with deep convolutional neural networks. Communications of the ACM, 60(6), 84–90. https://doi.org/10.1145/3065386
LeCun, Y., Bengio, Y., & Hinton, G. (2015). Deep learning. Nature, 521(7553), 436–444. https://doi.org/10.1038/nature14539
Li, X., & Kong, A. W. K. (2017). A multi-model restoration algorithm for recovering blood vessels in skin images. Image and Vision Computing, 61, 40–53. https://doi.org/10.1016/j.imavis.2017.02.006
Lowes, M. A., Bowcock, A. M., & Krueger, J. G. (2007). Pathogenesis and therapy of psoriasis. Nature, 445(7130), 866–873. https://doi.org/10.1038/nature05663
Peng, L., Na, Y., Changsong, D., Sheng, L. I., & Hui, M. (2021). Research on classification diagnosis model of psoriasis based on deep residual network. Digital Chinese Medicine, 4(2), 92–101. https://doi.org/10.1016/j.dcmed.2021.06.003
Pham, T.-C., Doucet, A., Luong, C.-M., Tran, C.-T., & Hoang, V.-D. (2020). Improving Skin-Disease Classification Based on Customized Loss Function Combined With Balanced Mini-Batch Logic and Real-Time Image Augmentation. IEEE Access, 8, 150725–150737. https://doi.org/10.1109/ACCESS.2020.3016653
Sandler, M., Howard, A., Zhu, M., Zhmoginov, A., & Chen, L.-C. (2018). MobileNetV2: Inverted Residuals and Linear Bottlenecks. 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, 4510–4520. https://doi.org/10.1109/CVPR.2018.00474
Singh, A., Verma, N., & Shukla, S. (2023). Deep learning framework for psoriasis detection using VGG and Inception models.
Tan, M., & Le, Q. V. (2019). EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. 97, 6105–6114. https://doi.org/http://dx.doi.org/10.48550/arXiv.1905.11946
Wang, J., Zhang, Y., Xie, F., & Liu, J. (2025). Enhancing Diagnosis of Psoriasis and Inflammatory Skin Diseases: A Spatially Aligned Multimodal Model Integrating Clinical and Dermoscopic Images. Journal of Investigative Dermatology. https://doi.org/10.1016/j.jid.2025.03.034
Wijesinghe, L., Kulasekera, D., & Ilmini, W. (2019). An Intelligent Approach to Segmentation and Classification of Common Skin Diseases in Sri Lanka. 2019 National Information Technology Conference (NITC), 47–52. https://doi.org/10.1109/NITC48475.2019.9114507
Yang, Y., Wang, J., Xie, F., Liu, J., Shu, C., Wang, Y., Zheng, Y., & Zhang, H. (2021). A convolutional neural network trained with dermoscopic images of psoriasis performed on par with 230 dermatologists. Computers in Biology and Medicine, 139, 104924. https://doi.org/10.1016/j.compbiomed.2021.104924
Zhu, W., Lai, H., Zhang, H., Zhang, G., Luo, Y., Wang, J., Sun, L., Lu, J., Wang, S., & Xiang, Y. (2024). Abscissa-Ordinate Focused Network for Psoriasis and Eczema Healthcare Cyber-Physical System With Active Label Smoothing. IEEE Access, 12, 54953–54963. https://doi.org/10.1109/ACCESS.2024.3384310
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 Charu Bolia, Sunil Joshi

This work is licensed under a Creative Commons Attribution 4.0 International License.
The authors retain their rights:
a. The authors retain their trademark and patent rights, as well as any process or procedure described in the article.
b. The authors retain the right to share, copy, distribute, execute and publicly communicate the article published in the Revista Científica de Sistemas e Informática (RCSI) (for example, place it in an institutional repository or publish it in a book), with an acknowledgment of its initial publication in the RCSI.
c. Authors retain the right to make a subsequent publication of their work, to use the article or any part of it (for example: a compilation of their works, notes for conferences, thesis, or for a book), provided that they indicate the source of publication (authors of the work, journal, volume, number and date).








