Un marco de atención dual contrastiva para mejorar la extracción de relaciones entre eventos adversos de medicamentos
DOI:
https://doi.org/10.51252/rcsi.v5i2.968Palabras clave:
eventos adversos de medicamentos, PNL biomédica, aprendizaje contrastivo, atención dual, aprendizaje automático, procesamiento del lenguaje naturalResumen
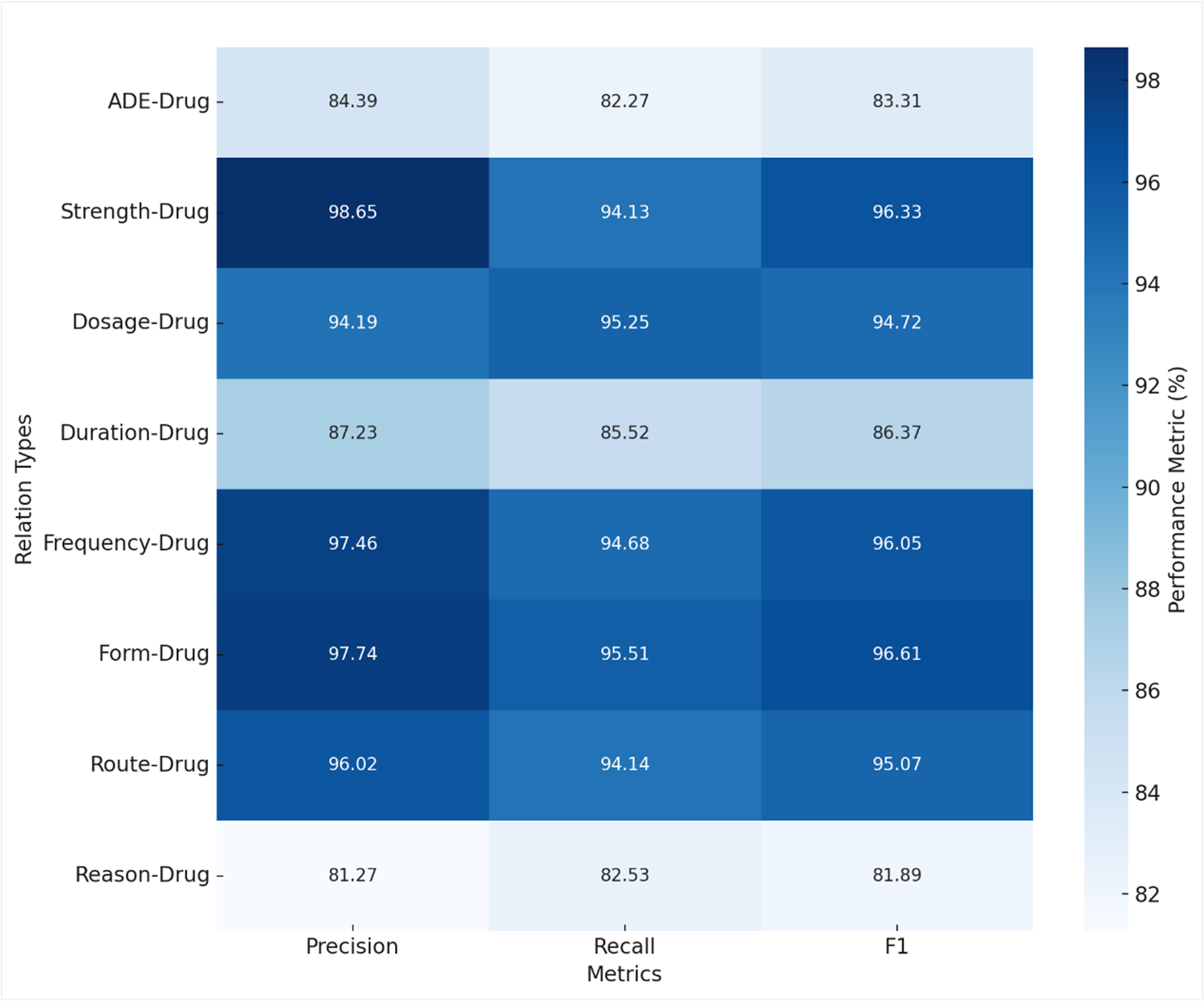
La extracción precisa de relaciones entre fármacos y eventos adversos a medicamentos (ADE) es fundamental para mejorar la seguridad del paciente. Sin embargo, los enfoques actuales tienen dificultades para captar relaciones complejas debido a limitaciones en la representación contextual. En el conjunto de datos n2c2, las instancias ADE-Fármaco (1107) son considerablemente menos numerosas que otras como Fuerza-Fármaco (6702) o Razón-Fármaco (5169), lo que introduce un fuerte desequilibrio que complica su identificación. Se emplea un modelo basado en codificadores de transformadores para generar representaciones contextuales, incorporando un mecanismo de atención dual que enfoca tanto en las entidades como en su entorno clínico. A través del aprendizaje contrastivo, se refina la representación de los pares de entidades, diferenciando con mayor precisión las relaciones correctas de las incorrectas. En las evaluaciones experimentales, se alcanzó un F1 general del 93,31 % y un 83,31 % en la relación ADE-Fármaco, superando a métodos previos. La combinación de codificación contextual, atención especializada y discriminación contrastiva permite afrontar con mayor eficacia los desafíos derivados del desequilibrio de clases y de la complejidad semántica del lenguaje clínico.
Descargas
Citas
Aguilar-Canto, F., Cardoso-Moreno, M., Jiménez, D., & Calvo, H. (2023). GPT-2 versus GPT-3 and Bloom: LLMs for LLMs Generative Text Detection. CEUR Workshop Proceedings, 3496.
Alsentzer, E., Murphy, J. R., Boag, W., Weng, W.-H., Jin, D., Naumann, T., & McDermott, M. B. A. (2019). Publicly Available Clinical BERT Embeddings. http://arxiv.org/abs/1904.03323
Brown, T. B., Mann, B., Ryder, N., Subbiah, M., Kaplan, J., Dhariwal, P., Neelakantan, A., Shyam, P., Sastry, G., Askell, A., Agarwal, S., Herbert-Voss, A., Krueger, G., Henighan, T., Child, R., Ramesh, A., Ziegler, D. M., Wu, J., Winter, C., … Amodei, D. (2020). Language Models are Few-Shot Learners. http://arxiv.org/abs/2005.14165
Christopoulou, F., Tran, T. T., Sahu, S. K., Miwa, M., & Ananiadou, S. (2020). Adverse drug events and medication relation extraction in electronic health records with ensemble deep learning methods. Journal of the American Medical Informatics Association, 27(1), 39-46. https://doi.org/10.1093/jamia/ocz101
Devlin, J., Chang, M.-W., Lee, K., & Toutanova, K. (2018). BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. http://arxiv.org/abs/1810.04805
El-allaly, E., Sarrouti, M., En-Nahnahi, N., & Ouatik El Alaoui, S. (2021). MTTLADE: A multi-task transfer learning-based method for adverse drug events extraction. Information Processing & Management, 58(3), 102473. https://doi.org/10.1016/j.ipm.2020.102473
Gao, T., Yao, X., & Chen, D. (2021). SimCSE: Simple Contrastive Learning of Sentence Embeddings. http://arxiv.org/abs/2104.08821
Gurulingappa, H., Mateen‐Rajpu, A., & Toldo, L. (2012). Extraction of potential adverse drug events from medical case reports. Journal of Biomedical Semantics, 3(1), 15. https://doi.org/10.1186/2041-1480-3-15
IOM. (2007). Preventing Medication Errors: Quality Chasm Series. En The National Academies Press, Washington, DC. Institute of Medicine.
Ji, Z., Wei, Q., & Xu, H. (2019). BERT-based Ranking for Biomedical Entity Normalization. http://arxiv.org/abs/1908.03548
Johnson, A. E. W., Bulgarelli, L., & Pollard, T. J. (2020). Deidentification of free-text medical records using pre-trained bidirectional transformers. Proceedings of the ACM Conference on Health, Inference, and Learning, 214-221. https://doi.org/10.1145/3368555.3384455
Johnson, A. E. W., Pollard, T. J., Shen, L., Lehman, L. H., Feng, M., Ghassemi, M., Moody, B., Szolovits, P., Anthony Celi, L., & Mark, R. G. (2016). MIMIC-III, a freely accessible critical care database. Scientific Data, 3(1), 160035. https://doi.org/10.1038/sdata.2016.35
Kim, Y. H., Kim, C., & Kim, Y. S. (2024). Language Model-Based Text Augmentation System for Cerebrovascular Disease-Related Medical Reports. Applied Sciences, 14(19).
Kim, Y., & Meystre, S. M. (2020). Ensemble method–based extraction of medication and related information from clinical texts. Journal of the American Medical Informatics Association, 27(1), 31-38. https://doi.org/10.1093/jamia/ocz100
Kumar, A., Chaudhary, K., Singh, P., & Thapa, J. (2020). Contextual Data Augmentation for Improving Natural Language Understanding in Biomedical Domain. IEEE BIBM.
Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H., & Kang, J. (2019). BioBERT: a pre-trained biomedical language representation model for biomedical text mining. https://doi.org/10.1093/bioinformatics/btz682
Li, Y., Tao, W., Li, Z., Sun, Z., Li, F., Fenton, S., Xu, H., & Tao, C. (2024). Artificial intelligence-powered pharmacovigilance: A review of machine and deep learning in clinical text-based adverse drug event detection for benchmark datasets. Journal of Biomedical Informatics, 152, 104621. https://doi.org/10.1016/j.jbi.2024.104621
Liu, S., Tang, B., Chen, Q., & Wang, X. (2016). Drug-Drug Interaction Extraction via Convolutional Neural Networks. Computational and Mathematical Methods in Medicine, 2016, 1-8. https://doi.org/10.1155/2016/6918381
Modi, S., Kasmiran, K. A., Mohd Sharef, N., & Sharum, M. Y. (2024). Extracting adverse drug events from clinical Notes: A systematic review of approaches used. Journal of Biomedical Informatics, 151, 104603. https://doi.org/10.1016/j.jbi.2024.104603
Murphy, S. N., Weber, G., Mendis, M., Gainer, V., Chueh, H. C., Churchill, S., & Kohane, I. (2010). Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). Journal of the American Medical Informatics Association, 17(2), 124-130. https://doi.org/10.1136/jamia.2009.000893
Peng, Y., Yan, S., & Lu, Z. (2018). Transfer Learning in Biomedical Natural Language Processing: An Evaluation of BERT and ELMo on Ten Benchmarking Datasets. Proceedings of the 18th BioNLP Workshop and Shared Task, 58-65. https://doi.org/10.18653/v1/W19-5006
Peng, Y., Yan, S., & Lu, Z. (2020). Biomedical Sentence Similarity Estimation with Transfer Learning and Entity-aware Contrastive Learning. BioNLP Workshop - ACL.
Roberts, K., Demner-Fushman, D., & Tonellato, P. J. (2009). Identifying and classifying adverse drug events in discharge summaries. AMIA Annual Symposium Proceedings, 601–605.
Sun, M., Zhang, Z., Han, X., Liu, Z., Jiang, X., & Liu, Q. (2019). ERNIE: Enhanced Language Representation with Informative Entities. http://arxiv.org/abs/1905.07129
Thapa, S., Islamaj, R., Rogers, K., & Demner-Fushman, D. (2022). Transfer Learning and Domain Adaptation with Pre-trained Embeddings for Clinical Natural Language Processing: A Case Study on Adverse Drug Event Classification. Journal of the American Medical Informatics Association, 29(6), 1019–1027. https://academic.oup.com/jamia/search-results?page=1&q=TransferLearningandDomainAdaptationwithPre-trainedEmbeddingsforClinicalNaturalLanguageProcessing%3AACaseStudyonAdverseDrugEventClassification&fl_SiteID=5396&SearchSourceType=1&allJ
Vig, J. (2019a). A Multiscale Visualization of Attention in the Transformer Model. http://arxiv.org/abs/1906.05714
Vig, J. (2019b). Visualizing Attention in Transformer-Based Language Representation Models. http://arxiv.org/abs/1904.02679
Wei, Q., Ji, Z., Li, Z., Du, J., Wang, J., Xu, J., Xiang, Y., Tiryaki, F., Wu, S., Zhang, Y., Tao, C., & Xu, H. (2020). A study of deep learning approaches for medication and adverse drug event extraction from clinical text. Journal of the American Medical Informatics Association, 27(1), 13-21. https://doi.org/10.1093/jamia/ocz063
WHO. (2008). The importance of pharmacovigilance - Safety Monitoring of medicinal products. World Health Organization.
Yang, X., Bian, J., Fang, R., Bjarnadottir, R. I., Hogan, W. R., & Wu, Y. (2020). Identifying relations of medications with adverse drug events using recurrent convolutional neural networks and gradient boosting. Journal of the American Medical Informatics Association, 27(1), 65-72. https://doi.org/10.1093/jamia/ocz144
Zhang, F., Zheng, W., Yu, J., & Qin, Y. (2020). Dual Attention-Based BiLSTM-CNN for Named Entity Recognition.
Zhang, Y., Chen, Q., Yang, Z., Lin, H., & Lu, Z. (2019). BioWordVec, improving biomedical word embeddings with subword information and MeSH. Scientific Data, 6(1), 52. https://doi.org/10.1038/s41597-019-0055-0
Zhang, Y., Sun, A., Jin, F., & Zhou, X. (2020). Contrastive learning for text classification. AAAI. http://arxiv.org/abs/2305.09269
Zhou, P., Shi, W., Tian, J., Qi, Z., Li, B., Hao, H., & Xu, B. (2016). Attention-Based Bidirectional Long Short-Term Memory Networks for Relation Classification. Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 2: Short Papers), 207-212. https://doi.org/10.18653/v1/P16-2034
Descargas
Publicado
Cómo citar
Número
Sección
Licencia
Derechos de autor 2025 Poonam Kashtriya, Pardeep Singh

Esta obra está bajo una licencia internacional Creative Commons Atribución 4.0.
Los autores retienen sus derechos:
a. Los autores retienen sus derechos de marca y patente, y tambien sobre cualquier proceso o procedimiento descrito en el artículo.
b. Los autores retienen el derecho de compartir, copiar, distribuir, ejecutar y comunicar públicamente el articulo publicado en la Revista Científica de Sistemas e Informática (RCSI) (por ejemplo, colocarlo en un repositorio institucional o publicarlo en un libro), con un reconocimiento de su publicación inicial en la RCSI.
c. Los autores retienen el derecho a hacer una posterior publicación de su trabajo, de utilizar el artículo o cualquier parte de aquel (por ejemplo: una compilación de sus trabajos, notas para conferencias, tesis, o para un libro), siempre que indiquen la fuente de publicación (autores del trabajo, revista, volumen, número y fecha).









