Delineando las tendencias temáticas en torno a la investigación de métodos computacionales en ciencias de la salud
DOI:
https://doi.org/10.51252/rcsi.v5i1.913Palabras clave:
métodos computacionales, ciencias de la salud, informática médica, análisis de coocurrencia de términos, bibliometríaResumen
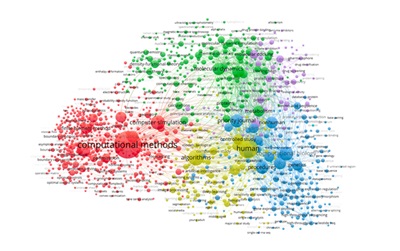
En este estudio, adoptamos el término método de análisis de coocurrencia para explorar las tendencias temáticas en la literatura sobre métodos computacionales aplicados a las ciencias de la salud. Esbozamos los temas y subtemas clave que caracterizan esta área de investigación. Se utilizó el algoritmo Visualization of Similarities (VOS) para representar las relaciones entre palabras clave en sus grupos temáticos. Se detectaron cinco grupos: métodos computacionales y modelización, química computacional y dinámica molecular, biología computacional y análisis de datos, métodos computacionales y modelos biológicos, y modelización computacional y diseño de fármacos. El estudio indica que la integración de métodos computacionales en las ciencias de la salud es un campo en continua expansión. Las aplicaciones clave incluyen la modelización de procesos biológicos, la simulación de interacciones moleculares y la optimización de tratamientos médicos. Los diversos grupos analizados demuestran que las herramientas computacionales mejoran la exploración de los fenómenos biomédicos y aumentan la precisión de los diagnósticos, la personalización de las terapias y la eficiencia de la investigación farmacéutica.
Descargas
Citas
Angenent, S., Pichon, E., & Tannenbaum, A. (2006). Mathematical methods in medical image processing. Bulletin of the American mathematical society, 43(3), 365-396.
Bardini, R., & Di Carlo, S. (2024). Computational methods for biofabrication in tissue engineering and regenerative medicine-a literature review. Computational and Structural Biotechnology Journal, 23, 601-616. https://doi.org/10.1016/j.csbj.2023.12.035
Boccaccio, A., Ballini, A., Pappalettere, C., Tullo, D., Cantore, S., & Desiate, A. (2011). Finite element method (FEM), mechanobiology and biomimetic scaffolds in bone tissue engineering. International journal of biological sciences, 7(1), 112. https://doi.org/10.7150/ijbs.7.112
Bonafe, C. F. S., Bispo, J. A. C., & de Jesus, M. B. (2018). The polygonal model: A simple representation of biomolecules as a tool for teaching metabolism. Biochemistry and Molecular Biology Education, 46(1), 66-75. https://doi.org/10.1002/bmb.21093
Cheng, L., Zhao, H., Wang, P., Zhou, W., Luo, M., Li, T., ... & Jiang, Q. (2019). Computational methods for identifying similar diseases. Molecular Therapy-Nucleic Acids, 18, 590-604. https://doi.org/10.1016/j.omtn.2019.09.019
Day, R. S. (2006). Challenges of biological realism and validation in simulation-based medical education. Artificial Intelligence in Medicine, 38(1), 47-66. https://doi.org/10.1016/j.artmed.2006.01.001
Faust, O. (2018). Documenting and predicting topic changes in Computers in Biology and Medicine: A bibliometric keyword analysis from 1990 to 2017. Informatics in Medicine Unlocked, 11, 15-27. https://doi.org/10.1016/j.imu.2018.03.002
Ghassemi, M., Naumann, T., Schulam, P., Beam, A. L., Chen, I. Y., & Ranganath, R. (2020). A review of challenges and opportunities in machine learning for health. AMIA Summits on Translational Science Proceedings, 2020, 191. https://pmc.ncbi.nlm.nih.gov/articles/PMC7233077/
Giorgetti, A., Ruggerone, P., Pantano, S., & Carloni, P. (2012). Advanced computational methods in molecular medicine. Journal of Biomedicine and Biotechnology, 2012. https://doi.org/10.1155/2012/709085
Jiao, X., Jin, X., Ma, Y., Yang, Y., Li, J., Liang, L., ... & Li, Z. (2021). A comprehensive application: Molecular docking and network pharmacology for the prediction of bioactive constituents and elucidation of mechanisms of action in component-based Chinese medicine. Computational Biology and Chemistry, 90, 107402. https://doi.org/10.1016/j.compbiolchem.2020.107402
Krzywanski, J., Sosnowski, M., Grabowska, K., Zylka, A., Lasek, L., & Kijo-Kleczkowska, A. (2024). Advanced Computational Methods for Modeling, Prediction and Optimization—A Review. Materials, 17(14), 3521. https://doi.org/10.3390/ma17143521
Lin, H. (2022). Computational methods and resources in biological and medical data. Current Medicinal Chemistry, 29(5), 786-788. https://doi.org/10.2174/092986732905220214141331
Mathews, J. D., McCaw, C. T., McVernon, J., McBryde, E. S., & McCaw, J. M. (2007). A biological model for influenza transmission: pandemic planning implications of asymptomatic infection and immunity. PLoS One, 2(11), e1220. https://doi.org/10.1371/journal.pone.0001220
Mazurek, A. H., Szeleszczuk, Ł., & Pisklak, D. M. (2020). Periodic DFT calculations—review of applications in the pharmaceutical sciences. Pharmaceutics, 12(5), 415. https://doi.org/10.3390/pharmaceutics12050415
Oscuvilca Tapia, E. C., Albitres Infantes, J. J., Cadenas Calderón, P. C., Aguinaga Mendoza, G. M., Paredes Jiménez, H. R., & Andrade Girón, E. C. (2024). Health and medical informatics research: Identifying international collaboration patterns at the country and institution level. Iberoamerican Journal of Science Measurement and Communication, 4(3), 1–16. https://doi.org/10.47909/ijsmc.137
Piana, M. (2009). Computational Methods in Biomedical Imaging. In Encyclopedia of Artificial Intelligence (pp. 372-376). IGI Global. DOI: 10.4018/978-1-59904-849-9.ch057
Poiate, I. A., Vasconcellos, A. B., Mori, M., & Poiate Jr, E. (2011). 2D and 3D finite element analysis of central incisor generated by computerized tomography. Computer methods and programs in biomedicine, 104(2), 292-299. https://doi.org/10.1016/j.cmpb.2011.03.017
Quesne, M. G., Borowski, T., & de Visser, S. P. (2016). Quantum mechanics/molecular mechanics modeling of enzymatic processes: Caveats and breakthroughs. Chemistry–A European Journal, 22(8), 2562-2581. https://doi.org/10.1002/chem.201503802
Sheng, C., & Zhang, W. (2013). Fragment informatics and computational fragment‐based drug design: an overview and update. Medicinal Research Reviews, 33(3), 554-598. https://doi.org/10.1002/med.21255
Shukla, N., Merigó, J. M., Lammers, T., & Miranda, L. (2020). Half a century of computer methods and programs in biomedicine: A bibliometric analysis from 1970 to 2017. Computer methods and programs in biomedicine, 183, 105075. https://doi.org/10.1016/j.cmpb.2019.105075
Solorzano, G., & Plevris, V. (2022). Computational intelligence methods in simulation and modeling of structures: A state-of-the-art review using bibliometric maps. Frontiers in Built Environment, 8, 1049616. https://doi.org/10.3389/fbuil.2022.1049616
Sousa, S. F., Ribeiro, A. J., Neves, R. P., Brás, N. F., Cerqueira, N. M., Fernandes, P. A., & Ramos, M. J. (2017). Application of quantum mechanics/molecular mechanics methods in the study of enzymatic reaction mechanisms. Wiley Interdisciplinary Reviews: Computational Molecular Science, 7(2), e1281. https://doi.org/10.1002/wcms.1281
Tang, X. (2019). The role of artificial intelligence in medical imaging research. BJR| Open, 2(1), 20190031. https://doi.org/10.1259/bjro.20190031
Tiwari, G., Shukla, A., Singh, A., & Tiwari, R. (2024). Computer simulation for effective pharmaceutical kinetics and dynamics: a review. Current Computer-Aided Drug Design, 20(4), 325-340. https://doi.org/10.2174/1573409919666230228104901
Udupa, J. K. (2005). Display of 3D information in discrete 3D scenes produced by computerized tomography. Proceedings of the IEEE, 71(3), 420-431. https://doi.org/10.1109/PROC.1983.12599
Usyk, T. P., & McCulloch, A. D. (2003). Computational methods for soft tissue biomechanics. In Biomechanics of soft tissue in cardiovascular systems (pp. 273-342). Vienna: Springer Vienna. https://doi.org/10.1007/978-3-7091-2736-0_7
Vaishya, R., Gupta, B. M., Singh, Y., Bansal, M., & Vaish, A. (2024). Global research on type 1 diabetes: A bibliometric study during 2001-2022. Iberoamerican Journal of Science Measurement and Communication, 4(2), 1–14. https://doi.org/10.47909/ijsmc.78
Wang, J. (2008). Computational biology of genome expression and regulation—a review of microarray bioinformatics. Journal of Environmental Pathology, Toxicology and Oncology, 27(3). DOI: 10.1615/JEnvironPatholToxicolOncol.v27.i3.10
Wang, K., Huang, Y., Wang, Y., You, Q., & Wang, L. (2024). Recent advances from computer-aided drug design to artificial intelligence drug design. RSC Medicinal Chemistry, 15(12), 3978-4000. https://doi.org/10.1039/D4MD00522H
Wu, Y., Krishnan, S., & Ghoraani, B. (2022). Computational methods for physiological signal processing and data analysis. Computational and Mathematical Methods in Medicine, 2022(1), 9861801. https://doi.org/10.1155/2022/9861801
Descargas
Publicado
Cómo citar
Número
Sección
Licencia
Derechos de autor 2025 Elsa Carmen Oscuvilca-Tapia, Fredy Ruperto Bermejo-Sánchez, Miriam Milagros Noreña-Lucho, Efraín Ademar Estrada-Choque

Esta obra está bajo una licencia internacional Creative Commons Atribución 4.0.
Los autores retienen sus derechos:
a. Los autores retienen sus derechos de marca y patente, y tambien sobre cualquier proceso o procedimiento descrito en el artículo.
b. Los autores retienen el derecho de compartir, copiar, distribuir, ejecutar y comunicar públicamente el articulo publicado en la Revista Científica de Sistemas e Informática (RCSI) (por ejemplo, colocarlo en un repositorio institucional o publicarlo en un libro), con un reconocimiento de su publicación inicial en la RCSI.
c. Los autores retienen el derecho a hacer una posterior publicación de su trabajo, de utilizar el artículo o cualquier parte de aquel (por ejemplo: una compilación de sus trabajos, notas para conferencias, tesis, o para un libro), siempre que indiquen la fuente de publicación (autores del trabajo, revista, volumen, número y fecha).








