Framing thematic trends around computational methods research in health sciences
DOI:
https://doi.org/10.51252/rcsi.v5i1.913Keywords:
computational methods, health sciences, health informatics, term co-occurrence analysis, bibliometricsAbstract
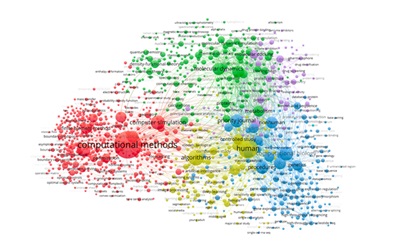
In this study, we adopted the term co-occurrence analysis method to explore the thematic trends in the literature regarding computational methods applied to health science. We will outline the key topics and subtopics that characterize this area of research. The Visualization of Similarities (VOS) algorithm was utilized to represent the relationships between keywords, identifying thematic clusters. Five clusters were identified: computational methods and modeling, computational chemistry and molecular dynamics, computational biology and data analysis, computational methods and biological models, and computational modeling and drug design. The study indicates that integrating computational methods in health sciences is a continuously expanding field. Key applications include modeling biological processes, simulating molecular interactions, and optimizing medical treatments. The various clusters analyzed demonstrate that computational tools enhance the exploration of biomedical phenomena and improve the accuracy of diagnoses, the personalization of therapies, and the efficiency of pharmaceutical research.
Downloads
References
Angenent, S., Pichon, E., & Tannenbaum, A. (2006). Mathematical methods in medical image processing. Bulletin of the American mathematical society, 43(3), 365-396.
Bardini, R., & Di Carlo, S. (2024). Computational methods for biofabrication in tissue engineering and regenerative medicine-a literature review. Computational and Structural Biotechnology Journal, 23, 601-616. https://doi.org/10.1016/j.csbj.2023.12.035
Boccaccio, A., Ballini, A., Pappalettere, C., Tullo, D., Cantore, S., & Desiate, A. (2011). Finite element method (FEM), mechanobiology and biomimetic scaffolds in bone tissue engineering. International journal of biological sciences, 7(1), 112. https://doi.org/10.7150/ijbs.7.112
Bonafe, C. F. S., Bispo, J. A. C., & de Jesus, M. B. (2018). The polygonal model: A simple representation of biomolecules as a tool for teaching metabolism. Biochemistry and Molecular Biology Education, 46(1), 66-75. https://doi.org/10.1002/bmb.21093
Cheng, L., Zhao, H., Wang, P., Zhou, W., Luo, M., Li, T., ... & Jiang, Q. (2019). Computational methods for identifying similar diseases. Molecular Therapy-Nucleic Acids, 18, 590-604. https://doi.org/10.1016/j.omtn.2019.09.019
Day, R. S. (2006). Challenges of biological realism and validation in simulation-based medical education. Artificial Intelligence in Medicine, 38(1), 47-66. https://doi.org/10.1016/j.artmed.2006.01.001
Faust, O. (2018). Documenting and predicting topic changes in Computers in Biology and Medicine: A bibliometric keyword analysis from 1990 to 2017. Informatics in Medicine Unlocked, 11, 15-27. https://doi.org/10.1016/j.imu.2018.03.002
Ghassemi, M., Naumann, T., Schulam, P., Beam, A. L., Chen, I. Y., & Ranganath, R. (2020). A review of challenges and opportunities in machine learning for health. AMIA Summits on Translational Science Proceedings, 2020, 191. https://pmc.ncbi.nlm.nih.gov/articles/PMC7233077/
Giorgetti, A., Ruggerone, P., Pantano, S., & Carloni, P. (2012). Advanced computational methods in molecular medicine. Journal of Biomedicine and Biotechnology, 2012. https://doi.org/10.1155/2012/709085
Jiao, X., Jin, X., Ma, Y., Yang, Y., Li, J., Liang, L., ... & Li, Z. (2021). A comprehensive application: Molecular docking and network pharmacology for the prediction of bioactive constituents and elucidation of mechanisms of action in component-based Chinese medicine. Computational Biology and Chemistry, 90, 107402. https://doi.org/10.1016/j.compbiolchem.2020.107402
Krzywanski, J., Sosnowski, M., Grabowska, K., Zylka, A., Lasek, L., & Kijo-Kleczkowska, A. (2024). Advanced Computational Methods for Modeling, Prediction and Optimization—A Review. Materials, 17(14), 3521. https://doi.org/10.3390/ma17143521
Lin, H. (2022). Computational methods and resources in biological and medical data. Current Medicinal Chemistry, 29(5), 786-788. https://doi.org/10.2174/092986732905220214141331
Mathews, J. D., McCaw, C. T., McVernon, J., McBryde, E. S., & McCaw, J. M. (2007). A biological model for influenza transmission: pandemic planning implications of asymptomatic infection and immunity. PLoS One, 2(11), e1220. https://doi.org/10.1371/journal.pone.0001220
Mazurek, A. H., Szeleszczuk, Ł., & Pisklak, D. M. (2020). Periodic DFT calculations—review of applications in the pharmaceutical sciences. Pharmaceutics, 12(5), 415. https://doi.org/10.3390/pharmaceutics12050415
Oscuvilca Tapia, E. C., Albitres Infantes, J. J., Cadenas Calderón, P. C., Aguinaga Mendoza, G. M., Paredes Jiménez, H. R., & Andrade Girón, E. C. (2024). Health and medical informatics research: Identifying international collaboration patterns at the country and institution level. Iberoamerican Journal of Science Measurement and Communication, 4(3), 1–16. https://doi.org/10.47909/ijsmc.137
Piana, M. (2009). Computational Methods in Biomedical Imaging. In Encyclopedia of Artificial Intelligence (pp. 372-376). IGI Global. DOI: 10.4018/978-1-59904-849-9.ch057
Poiate, I. A., Vasconcellos, A. B., Mori, M., & Poiate Jr, E. (2011). 2D and 3D finite element analysis of central incisor generated by computerized tomography. Computer methods and programs in biomedicine, 104(2), 292-299. https://doi.org/10.1016/j.cmpb.2011.03.017
Quesne, M. G., Borowski, T., & de Visser, S. P. (2016). Quantum mechanics/molecular mechanics modeling of enzymatic processes: Caveats and breakthroughs. Chemistry–A European Journal, 22(8), 2562-2581. https://doi.org/10.1002/chem.201503802
Sheng, C., & Zhang, W. (2013). Fragment informatics and computational fragment‐based drug design: an overview and update. Medicinal Research Reviews, 33(3), 554-598. https://doi.org/10.1002/med.21255
Shukla, N., Merigó, J. M., Lammers, T., & Miranda, L. (2020). Half a century of computer methods and programs in biomedicine: A bibliometric analysis from 1970 to 2017. Computer methods and programs in biomedicine, 183, 105075. https://doi.org/10.1016/j.cmpb.2019.105075
Solorzano, G., & Plevris, V. (2022). Computational intelligence methods in simulation and modeling of structures: A state-of-the-art review using bibliometric maps. Frontiers in Built Environment, 8, 1049616. https://doi.org/10.3389/fbuil.2022.1049616
Sousa, S. F., Ribeiro, A. J., Neves, R. P., Brás, N. F., Cerqueira, N. M., Fernandes, P. A., & Ramos, M. J. (2017). Application of quantum mechanics/molecular mechanics methods in the study of enzymatic reaction mechanisms. Wiley Interdisciplinary Reviews: Computational Molecular Science, 7(2), e1281. https://doi.org/10.1002/wcms.1281
Tang, X. (2019). The role of artificial intelligence in medical imaging research. BJR| Open, 2(1), 20190031. https://doi.org/10.1259/bjro.20190031
Tiwari, G., Shukla, A., Singh, A., & Tiwari, R. (2024). Computer simulation for effective pharmaceutical kinetics and dynamics: a review. Current Computer-Aided Drug Design, 20(4), 325-340. https://doi.org/10.2174/1573409919666230228104901
Udupa, J. K. (2005). Display of 3D information in discrete 3D scenes produced by computerized tomography. Proceedings of the IEEE, 71(3), 420-431. https://doi.org/10.1109/PROC.1983.12599
Usyk, T. P., & McCulloch, A. D. (2003). Computational methods for soft tissue biomechanics. In Biomechanics of soft tissue in cardiovascular systems (pp. 273-342). Vienna: Springer Vienna. https://doi.org/10.1007/978-3-7091-2736-0_7
Vaishya, R., Gupta, B. M., Singh, Y., Bansal, M., & Vaish, A. (2024). Global research on type 1 diabetes: A bibliometric study during 2001-2022. Iberoamerican Journal of Science Measurement and Communication, 4(2), 1–14. https://doi.org/10.47909/ijsmc.78
Wang, J. (2008). Computational biology of genome expression and regulation—a review of microarray bioinformatics. Journal of Environmental Pathology, Toxicology and Oncology, 27(3). DOI: 10.1615/JEnvironPatholToxicolOncol.v27.i3.10
Wang, K., Huang, Y., Wang, Y., You, Q., & Wang, L. (2024). Recent advances from computer-aided drug design to artificial intelligence drug design. RSC Medicinal Chemistry, 15(12), 3978-4000. https://doi.org/10.1039/D4MD00522H
Wu, Y., Krishnan, S., & Ghoraani, B. (2022). Computational methods for physiological signal processing and data analysis. Computational and Mathematical Methods in Medicine, 2022(1), 9861801. https://doi.org/10.1155/2022/9861801
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 Elsa Carmen Oscuvilca-Tapia, Fredy Ruperto Bermejo-Sánchez, Miriam Milagros Noreña-Lucho, Efraín Ademar Estrada-Choque

This work is licensed under a Creative Commons Attribution 4.0 International License.
The authors retain their rights:
a. The authors retain their trademark and patent rights, as well as any process or procedure described in the article.
b. The authors retain the right to share, copy, distribute, execute and publicly communicate the article published in the Revista Científica de Sistemas e Informática (RCSI) (for example, place it in an institutional repository or publish it in a book), with an acknowledgment of its initial publication in the RCSI.
c. Authors retain the right to make a subsequent publication of their work, to use the article or any part of it (for example: a compilation of their works, notes for conferences, thesis, or for a book), provided that they indicate the source of publication (authors of the work, journal, volume, number and date).








